Top Level Guide#
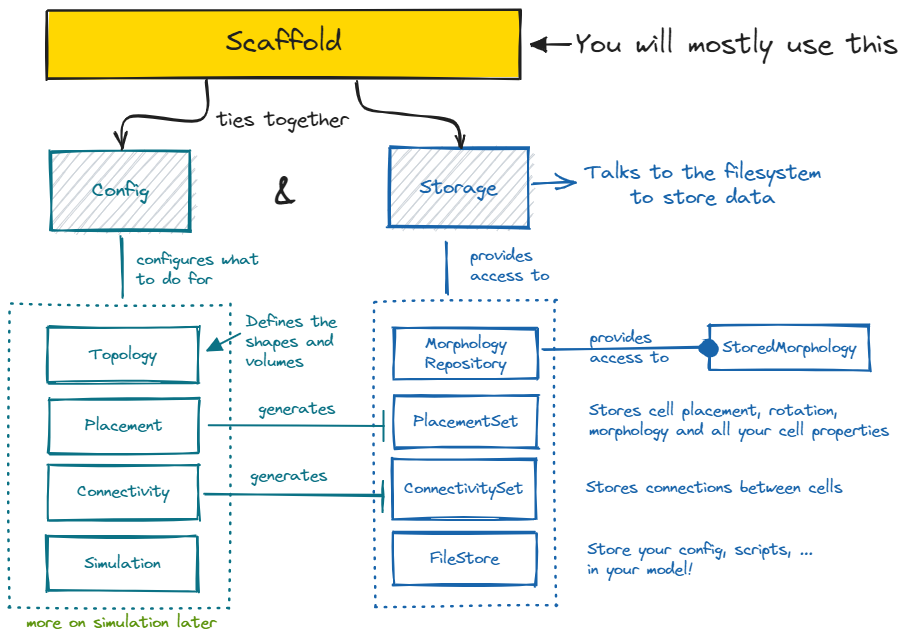
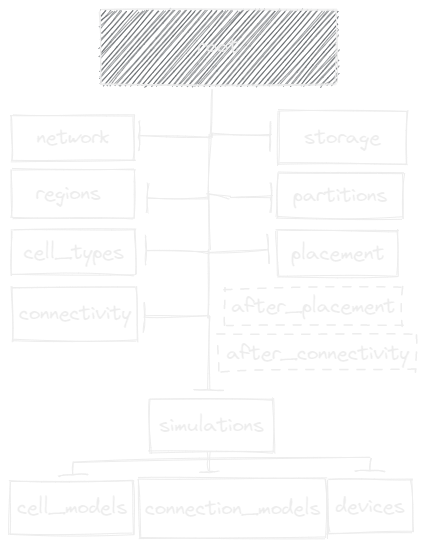
The Brain Scaffold Builder revolves around the Scaffold object. A
scaffold ties together all the information in the Configuration with the
Storage. The configuration contains your model description, while the
storage contains your model data, like concrete cell positions or connections.
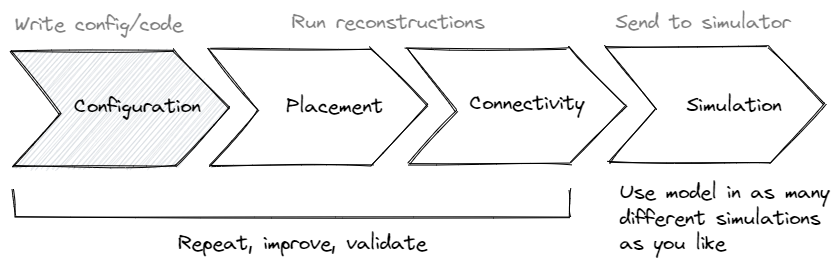
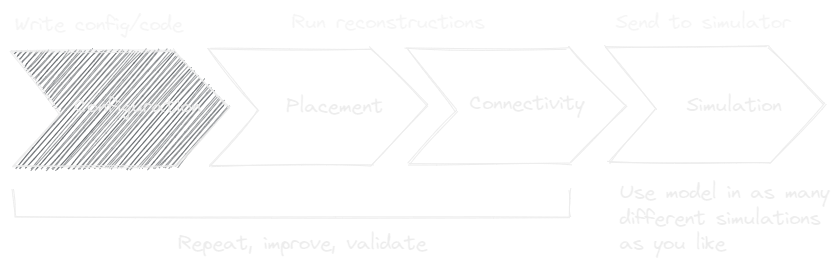
Using the scaffold object one can turn the abstract model configuration into a concrete
storage object full of neuroscience. For it to do so, the configuration needs to describe
which steps to take to place cells, called Placement, which steps to take to connect
cells, called Connectivity, and what representations to use during Simulation for
those cells and connections. All of these configurable objects can be accessed from the
scaffold object, under network.placement, network.connectivity,
network.simulations, …
Using the scaffold object, you can inspect the data in the storage by using the
PlacementSet and
ConnectivitySet APIs. PlacementSets can be obtained with
scaffold.get_placement_set, and
ConnectivitySets with scaffold.get_connectivity_set.
Ultimately this is the goal of the entire framework: To let you explicitly define every
component and parameter that is a part of your model, and all its parameters, in such a
way that a single CLI command, bsb compile, can turn your configuration into a
reconstructed biophysically detailed large scale neural network.
Workflow#
Configuration#