Morphologies#
Morphologies are the 3D representation of a cell. A morphology consists of head-to-tail connected branches, and branches consist of a series of points with radii. Points can be labelled and can have multiple user-defined properties per point.
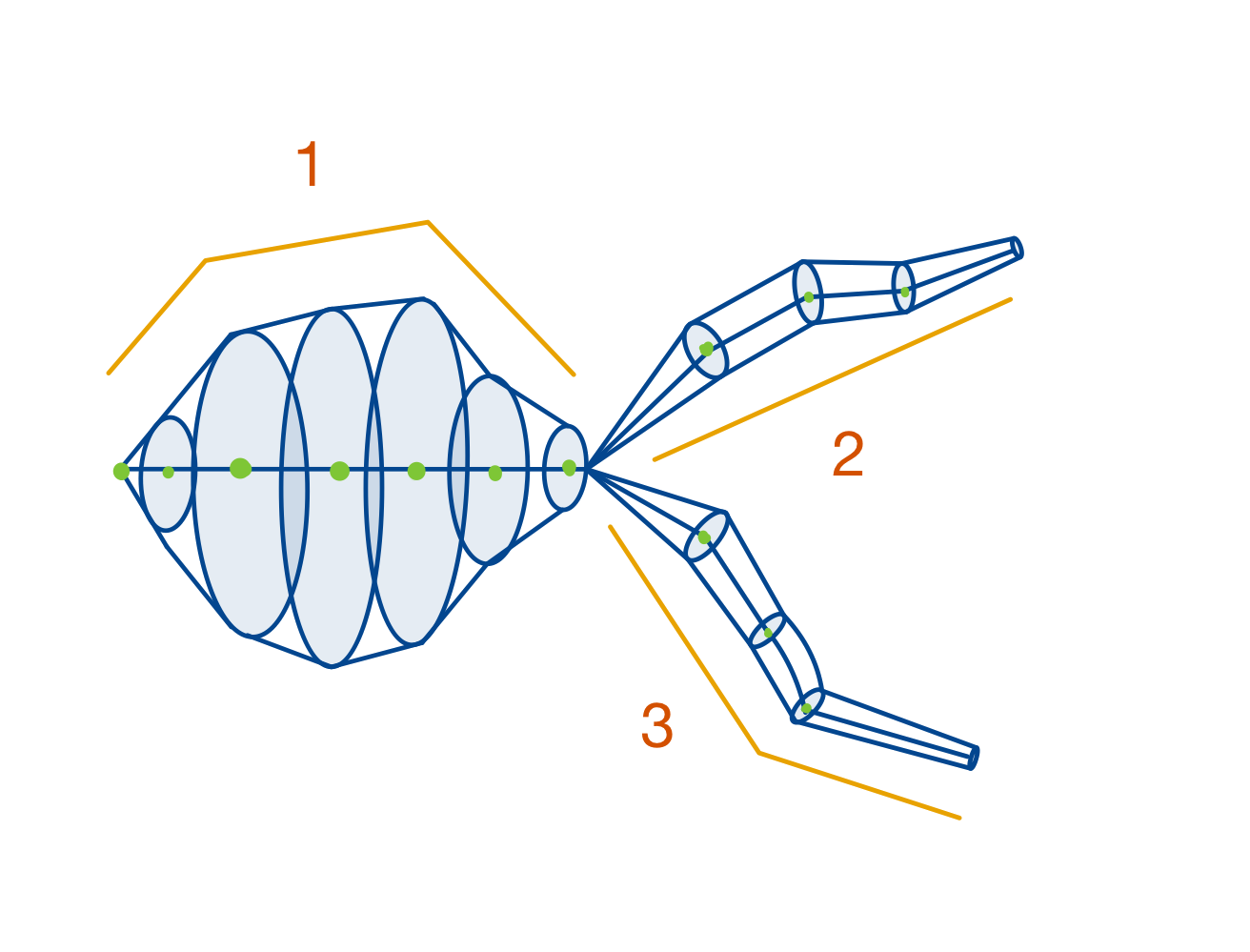
The root branch, shaped like a soma because of its radii.
A child branch of the root branch.
Another child branch of the root branch.
Network configurations can contain a morphologies key to define
the morphologies that should be processed and assigned to cells. See
Adding morphologies for a guide on the possibilities.
Morphologies can be stored in a network in the
MorphologyRepository.
Parsing morphologies#
A morphology file can be parsed with parse_morphology_file(),
if you already have the content of a file you can pass that directly into
parse_morphology_content():
from bsb import parse_morphology_file
morpho = parse_morphology_file("./my_file.swc")
Important
The default parser only supports SWC files. Use the morphio parser for ASC files.
There are many different formats and even multiple conventions per format for parsing morphologies. To support these diverse approaches the framework provides configurable Morphology parsers. You can pass the type of parser and additional arguments:
from bsb import parse_morphology_file
morpho = parse_morphology_file("./my_file.swc", parser="morphio", flags=["no_duplicates"])
Once we have our Morphology object we can save it in
Storage; storages and networks have a morphologies attribute that
links to a MorphologyRepository that can save and load
morphologies:
from bsb import Storage
store = Storage("hdf5", "morphologies.hdf5")
store.morphologies.save("MyCell", morpho)
Constructing morphologies#
Create your branches, attach them in a parent-child relationship, and provide the roots to
the Morphology constructor:
from bsb import Branch, Morphology
import numpy as np
root = Branch(
# XYZ
np.array([
[0, 1, 2],
[0, 1, 2],
[0, 1, 2],
]),
# radius
np.array([1, 1, 1]),
)
child_branch = Branch(
np.array([
[2, 3, 4],
[2, 3, 4],
[2, 3, 4],
]),
np.array([1, 1, 1]),
)
root.attach_child(child_branch)
m = Morphology([root])
Basic use#
Morphologies and branches contain spatial data in the points and radii attributes.
Points can be individually labelled with arbitrary strings, and additional properties for
each point can be assigned to morphologies/branches:
from bsb import from_storage
# Load the morphology
network = from_storage("network.hdf5")
morpho = network.morphologies.load("my_morphology")
print(f"Has {len(morpho)} points and {len(morpho.branches)} branches.")
Once loaded we can do transformations, label or assign properties on the morphology:
# Take a branch
special_branch = morpho.branches[3]
# Assign some labels to the whole branch
special_branch.label(["axon", "special"])
# Assign labels only to the first quarter of the branch
first_quarter = np.arange(len(special_branch)) < len(special_branch) / 4
special_branch.label(["initial_segment"], first_quarter)
# Assign random data as the `random_data` property to the branch
special_branch.set_property(random_data=np.random.random(len(special_branch)))
print(f"Random data for each point:", special_branch.random_data)
Once you’re done with the morphology you can save it again:
network.morphologies.save("processed_morphology", morpho)
Note
You can assign as many labels as you like (2^64 combinations max 😇)! Labels’ll cost you almost no memory or disk space! You can also add as many properties as you like, but they’ll cost you memory and disk space per point on the morphology.
Labels
Branches or points can be labelled, and pieces of the morphology can be selected by their label. Labels are also useful targets to insert biophysical mechanisms into parts of the cell later on in simulation.
import numpy as np
from bsb import from_storage
# Load the morphology
network = from_storage("network.hdf5")
morpho = network.morphologies.load("my_morphology")
# Filter branches
big_branches = [b for b in morpho.branches if np.any(b.radii > 2)]
for b in big_branches:
# Label all points on the branch as a `big_branch` point
b.label(["big_branch"])
if b.is_terminal:
# Label the last point on terminal branches as a `tip`
b.label(["tip"], [-1])
network.morphologies.save("labelled_morphology", morpho)
Properties
Branches and morphologies can be given additional properties. The basic properties are
x, y, z, radii and labels. You can pass additional properties to the
properties argument of the Branch constructor. They will be
automatically joined on the morphology.
Subtree transformations#
A subtree is a (sub)set of a morphology defined by a set of roots and all of its downstream branches (i.e. the branches emanating from a set of roots). A subtree with roots equal to the roots of the morphology is equal to the entire morphology, and all transformations valid on a subtree are also valid morphology transformations.
Creating subtrees#
Subtrees can be selected using label(s) on the morphology.
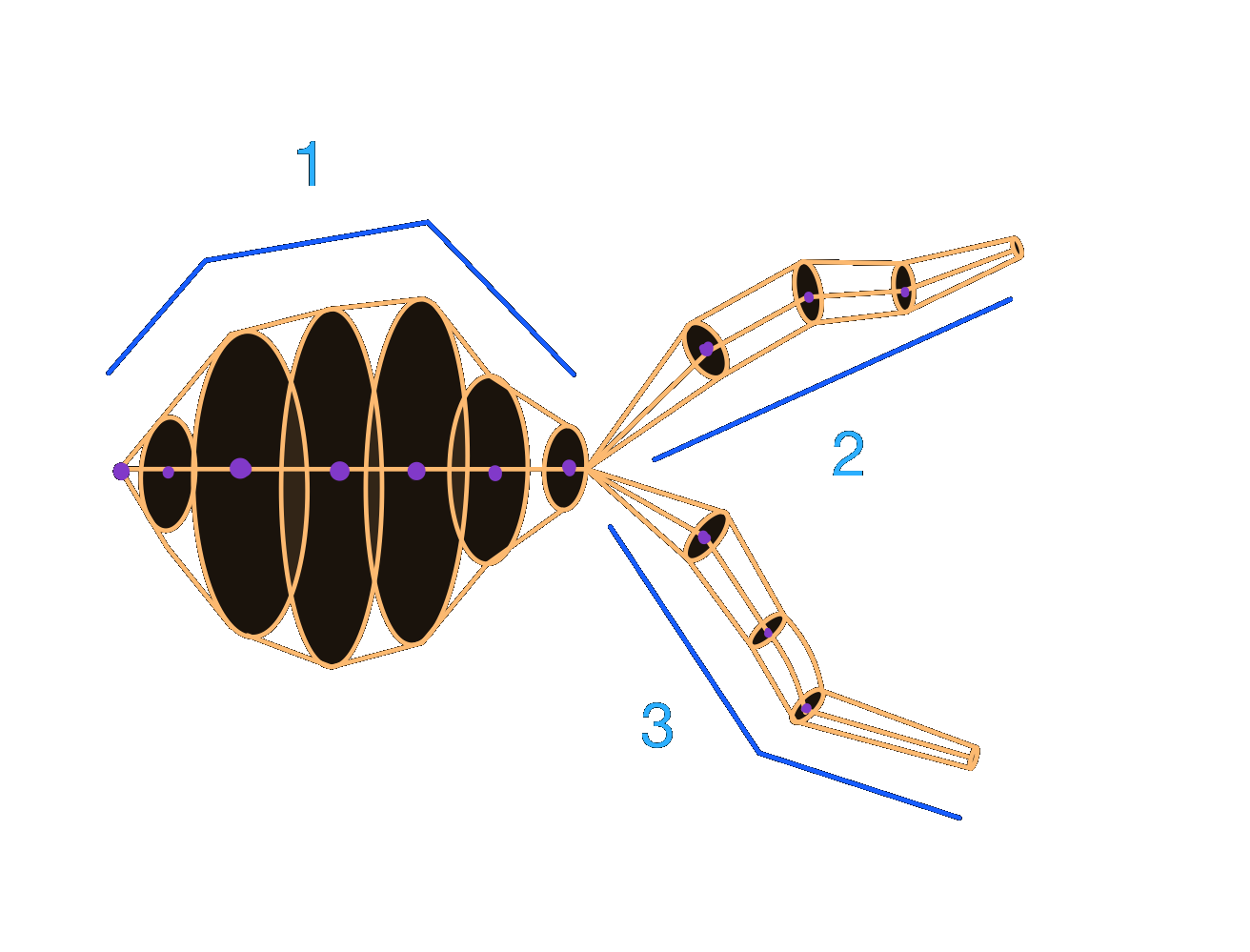
axon = morfo.subtree("axon")
# Multiple labels can be given
hybrid = morfo.subtree("proximal", "distal")
Warning
Branches will be selected as soon as they have one or more points labelled with a selected label.
Selections will always include all the branches emanating (downtree) from the selection as well:
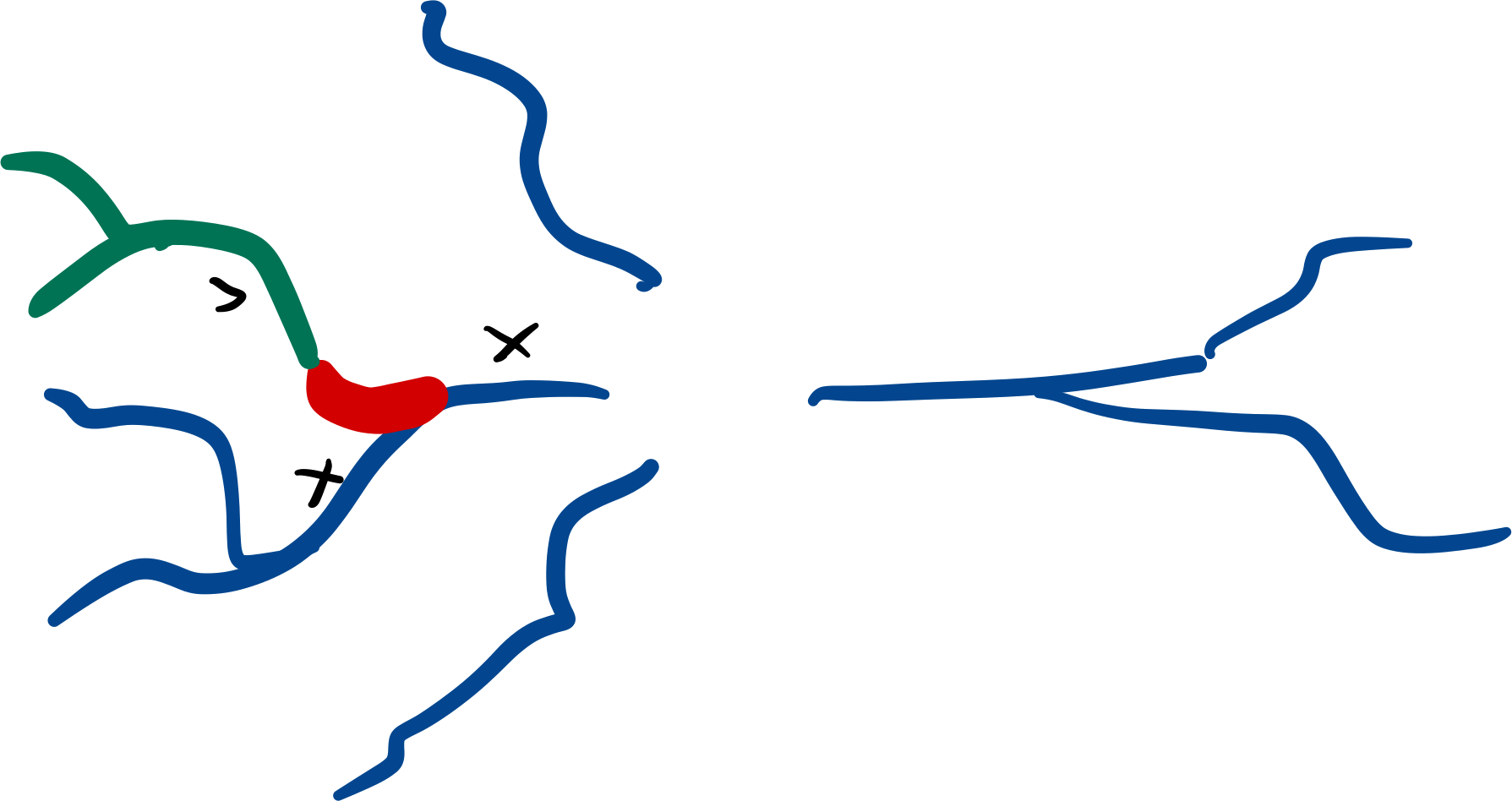
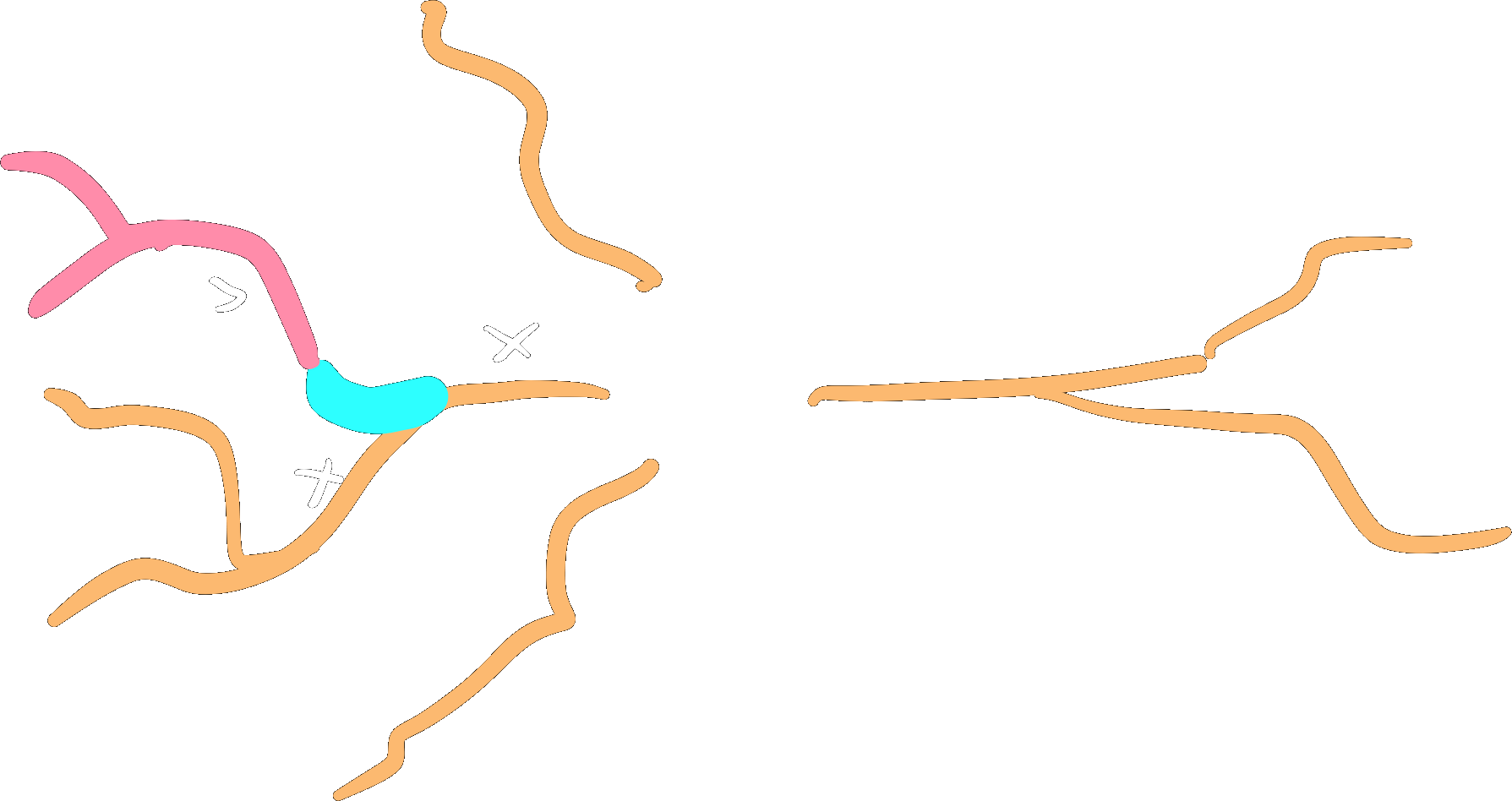
tuft = morfo.subtree("dendritic_piece")
Translation#
axon.translate([24, 100, 0])
Centering#
Subtrees may center() themselves so that the point (0, 0,
0) becomes the geometric mean of the roots.
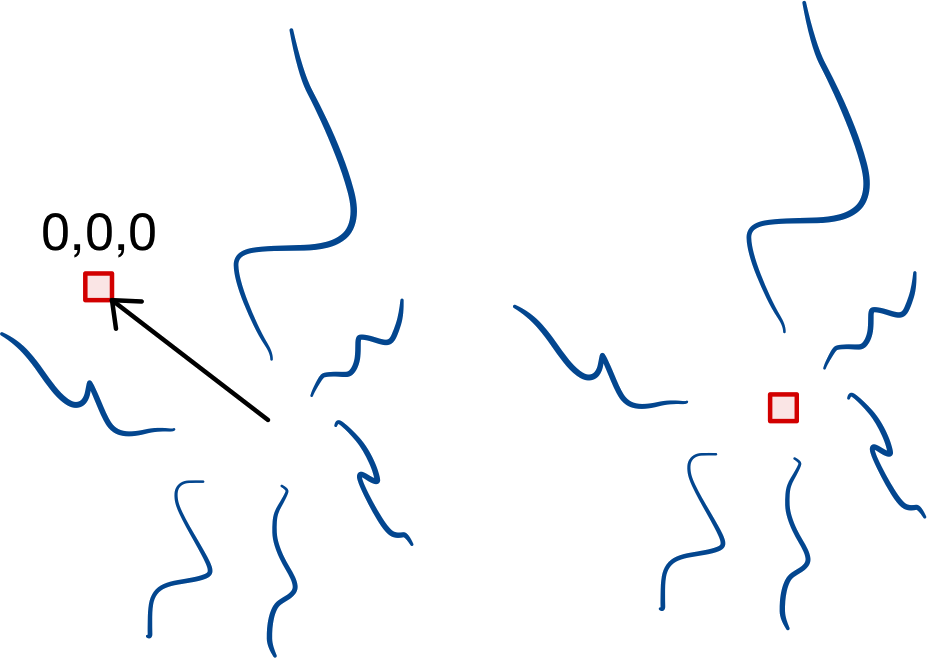
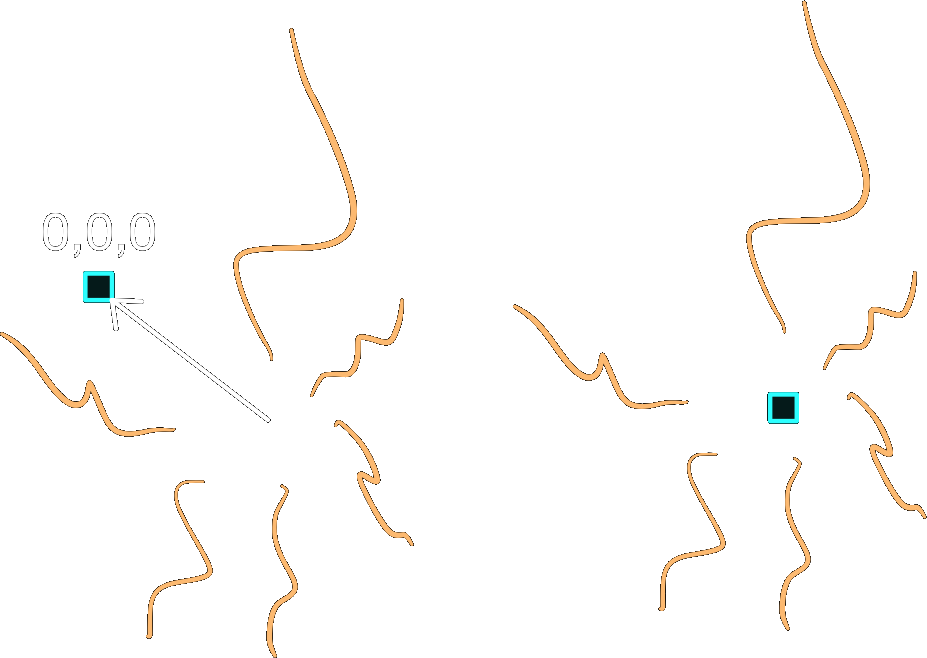
Rotation#
Subtrees may be rotated around a singular point, by
giving a Rotation (and a center, by default 0):
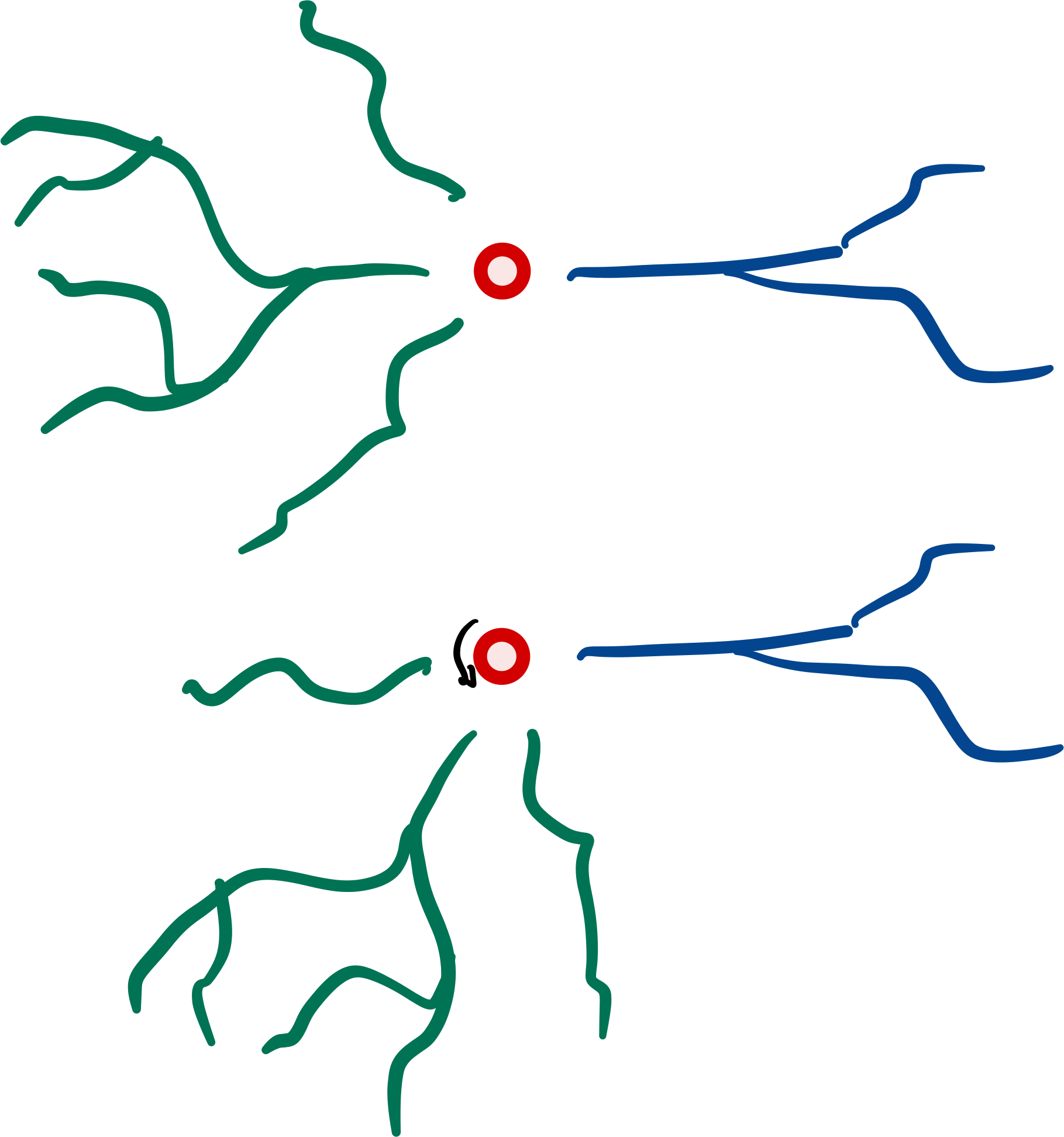
from scipy.spatial.transform import Rotation
r = Rotation.from_euler("xy", [90, 90], degrees=True)
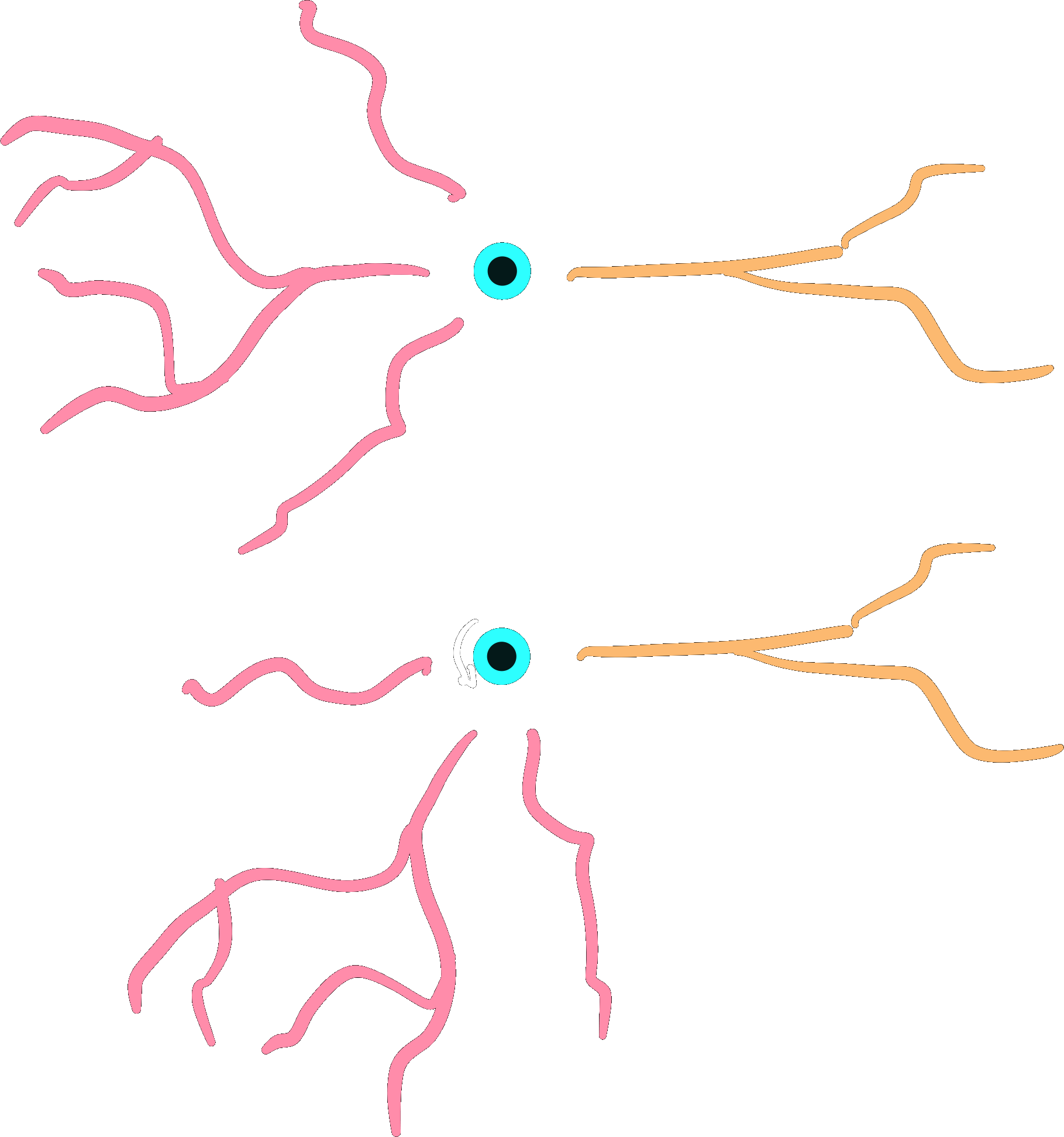
dendrites.rotate(r)
dendrite.rotate(r)
Note that this creates a gap, because we are rotating around the center, root-rotation might be preferred here.
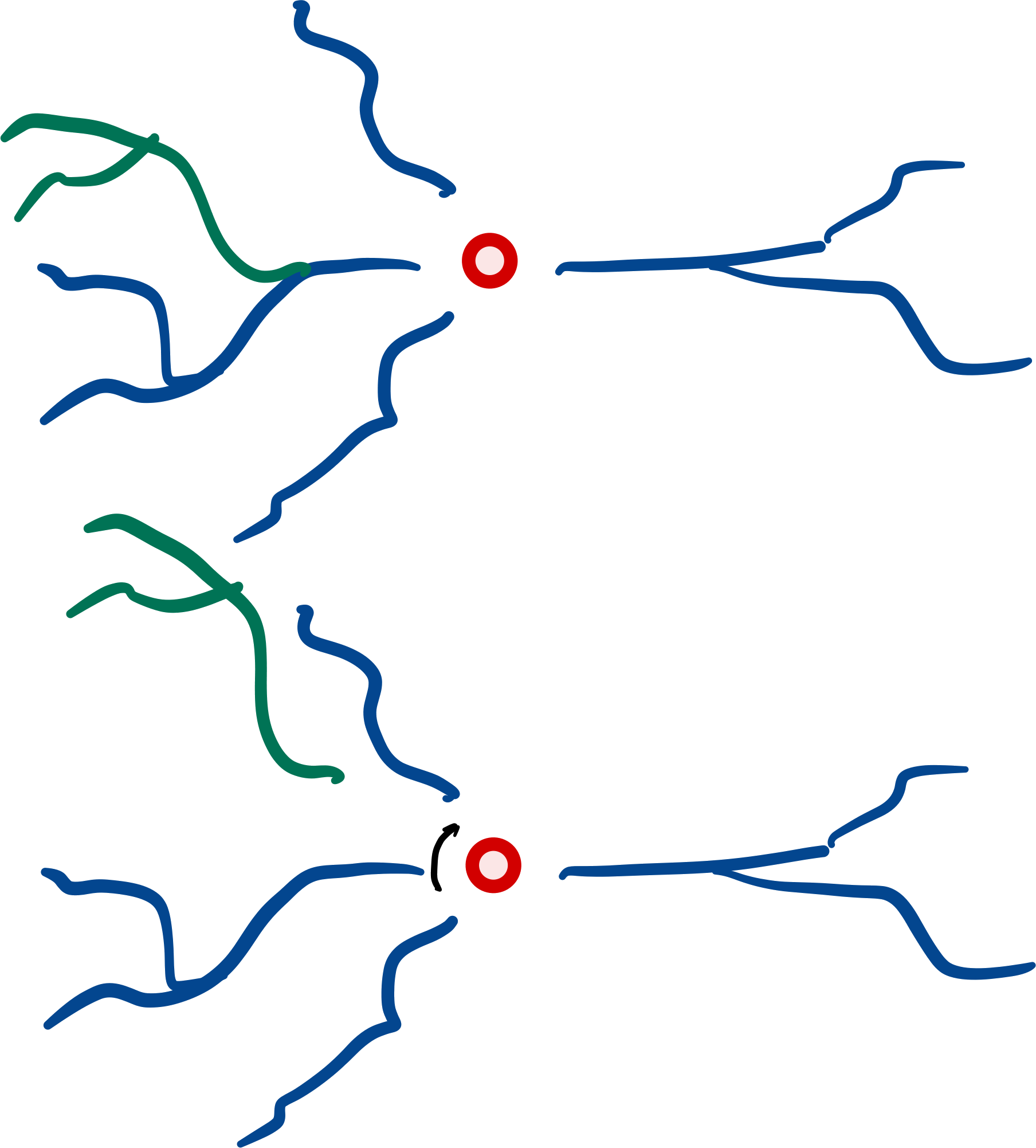
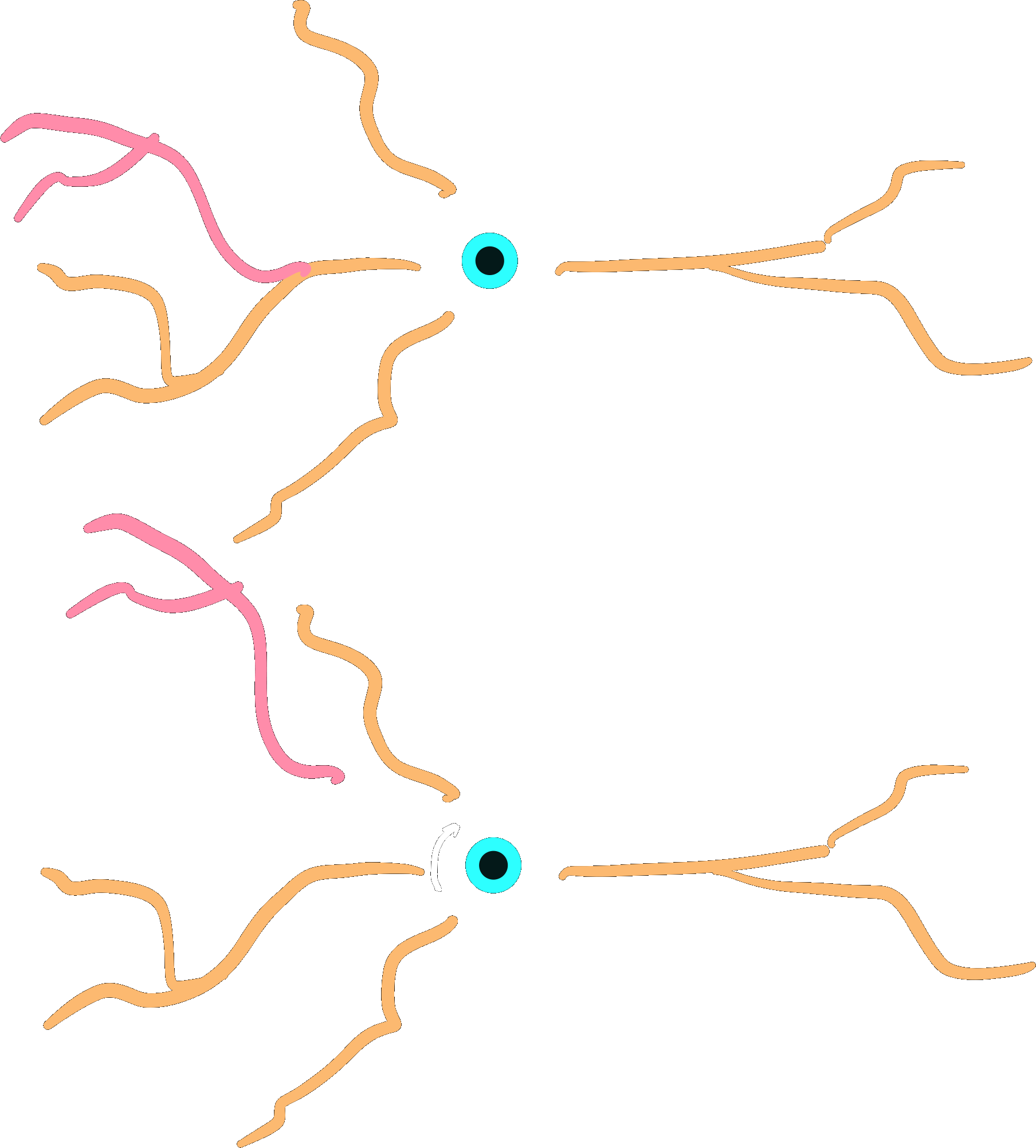
Root-rotation#
Subtrees may be root-rotated around each
respective root in the tree:
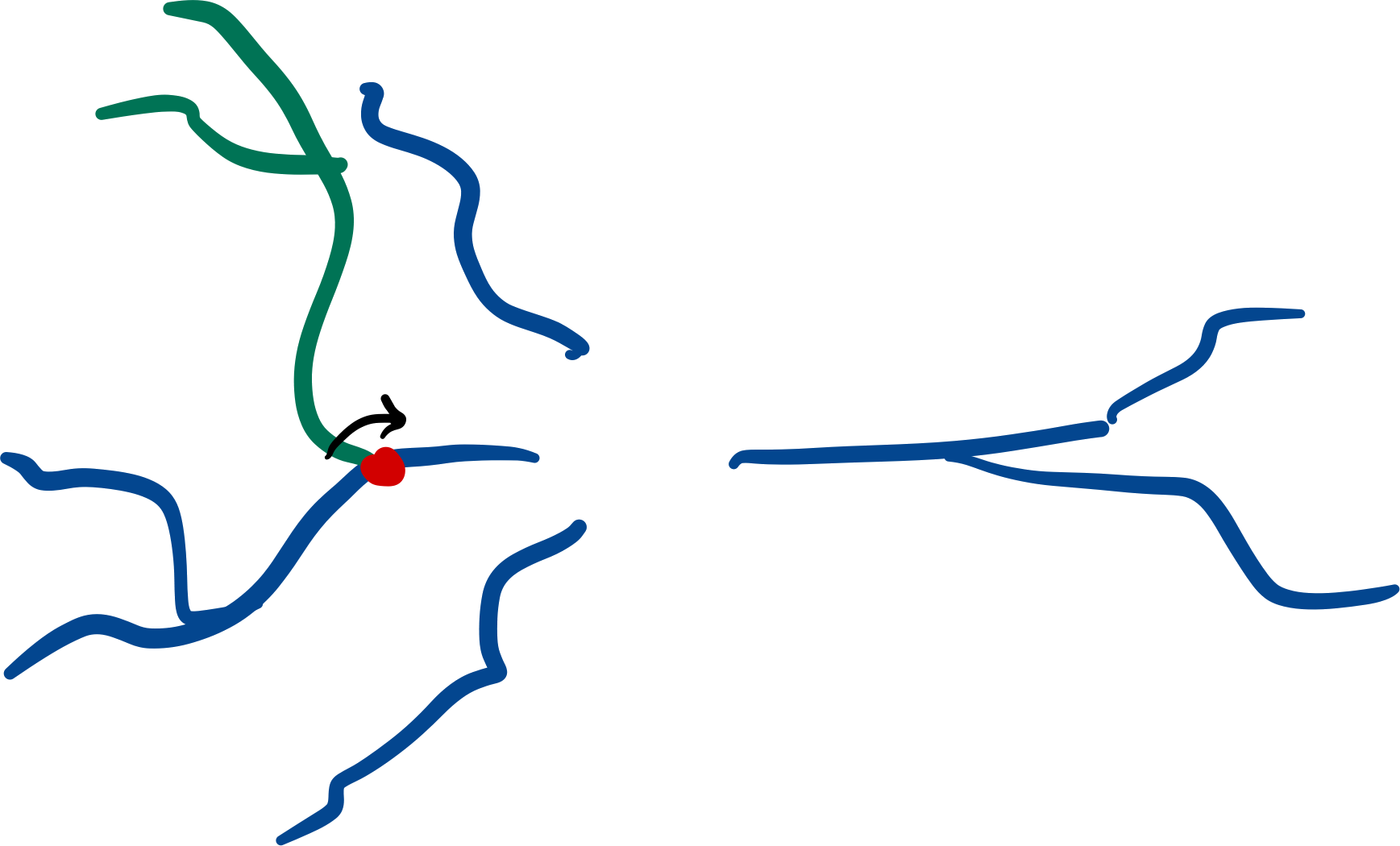
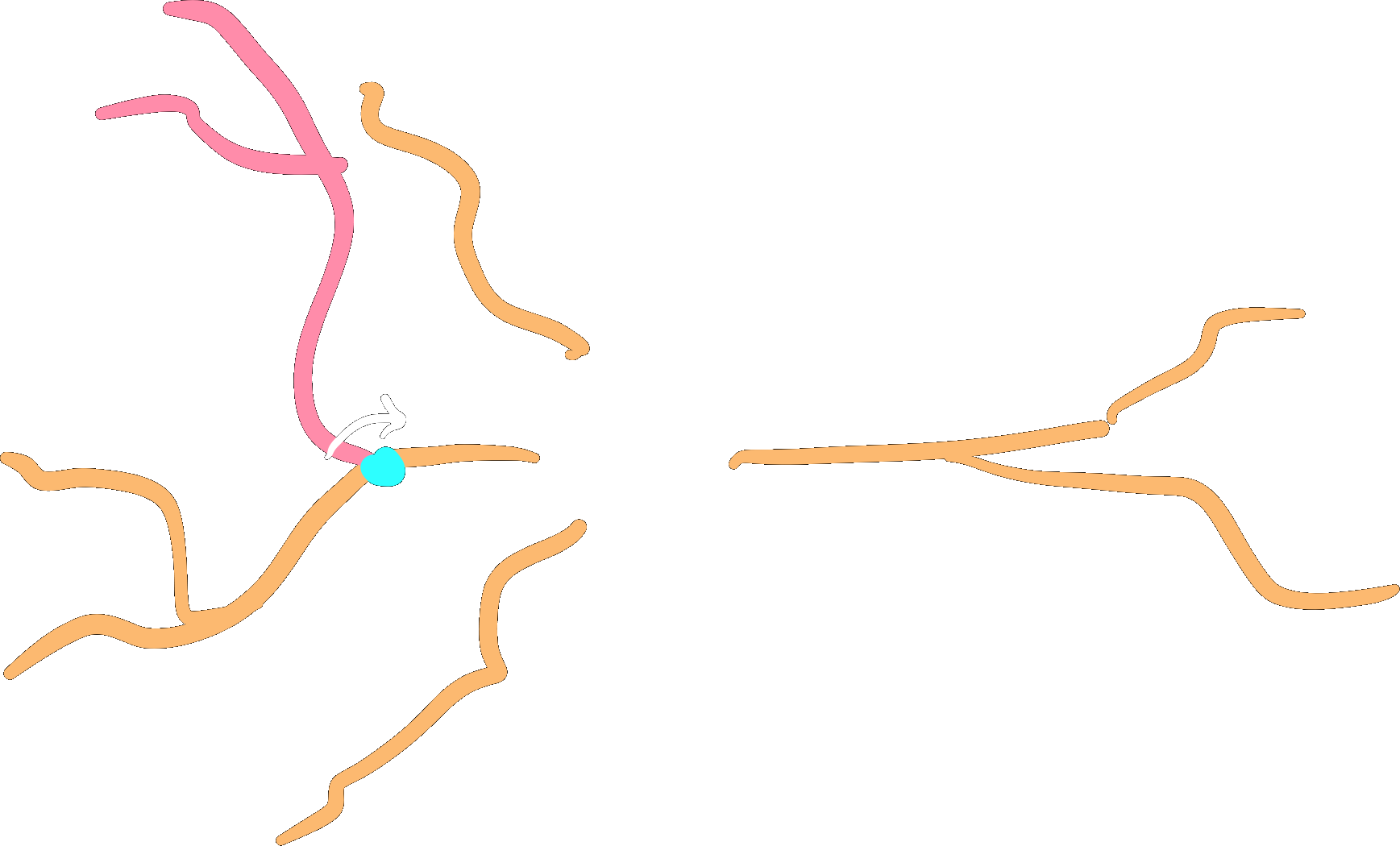
dendrite.root_rotate(r)
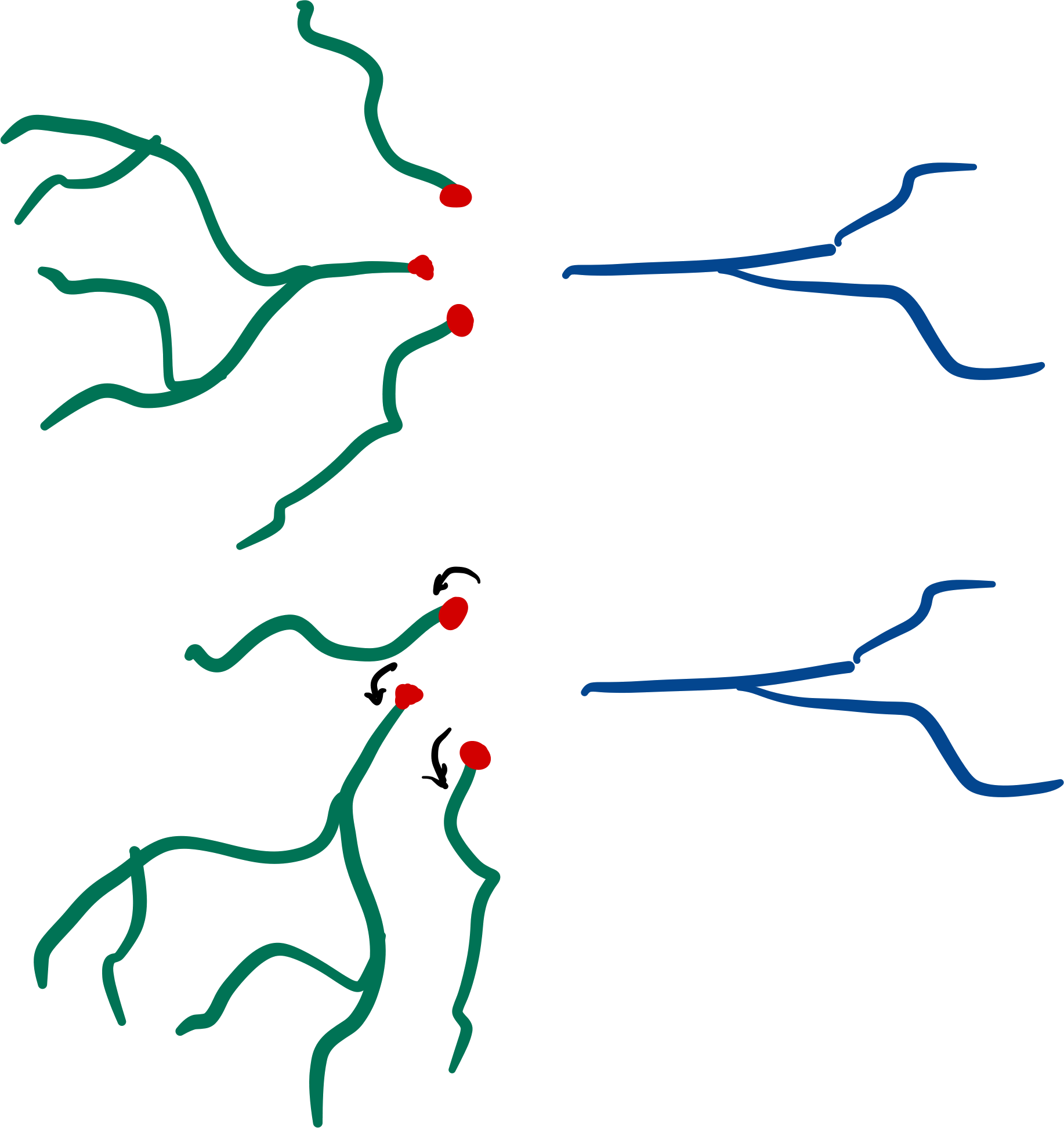
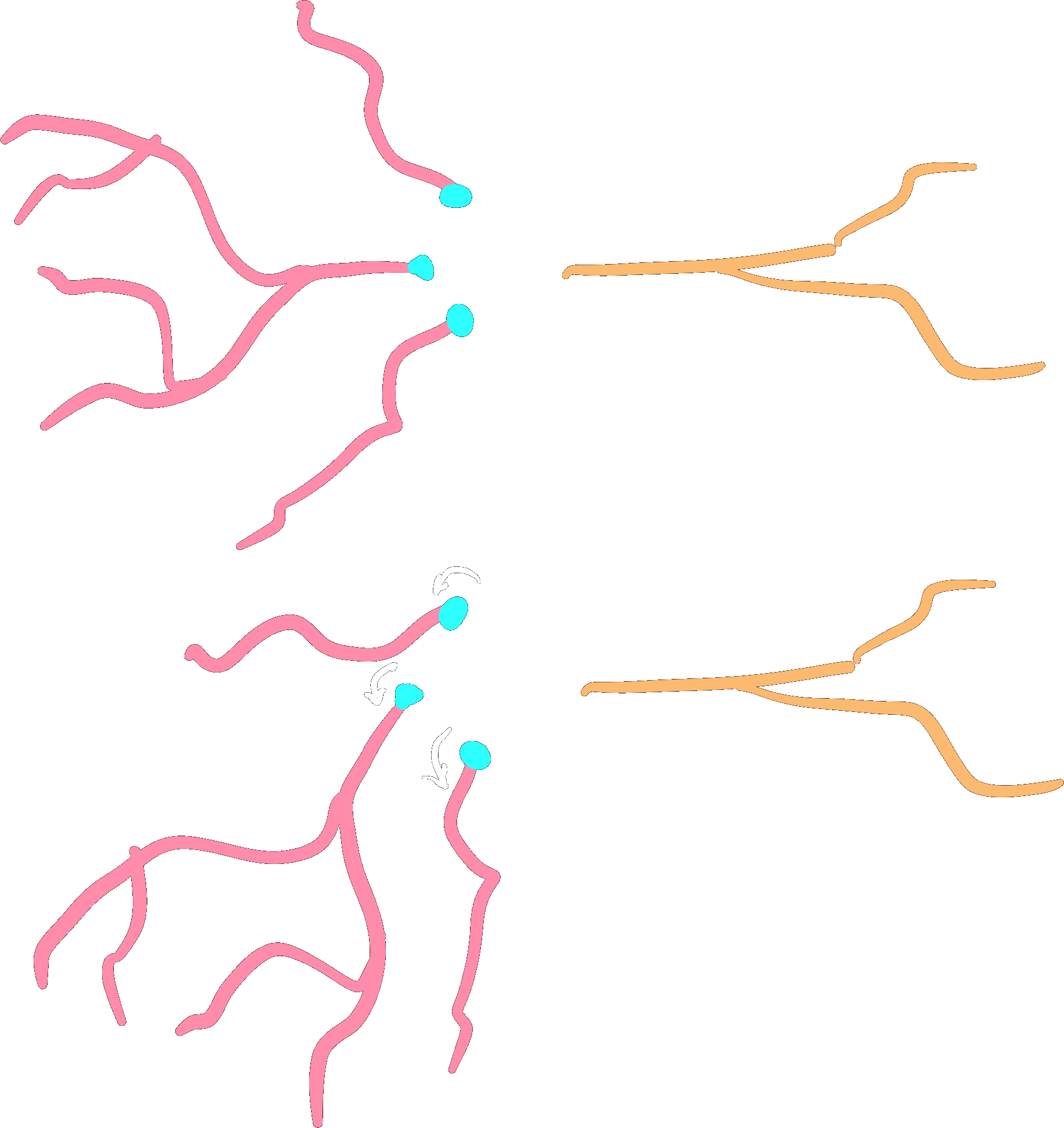
dendrites.root_rotate(r)
Additionally, you can root-rotate from a point of the
subtree instead of its root. In this case, points starting from the point selected will be rotated.
To do so, set the downstream_of parameter with the index of the point of your interest.
# rotate all points after the second point in the subtree
# i.e.: points at index 0 and 1 will not be rotated.
dendrites.root_rotate(r, downstream_of=2)
Note
This feature can only be applied to subtrees with a single root
Gap closing#
Subtree gaps between parent and child branches can be closed:
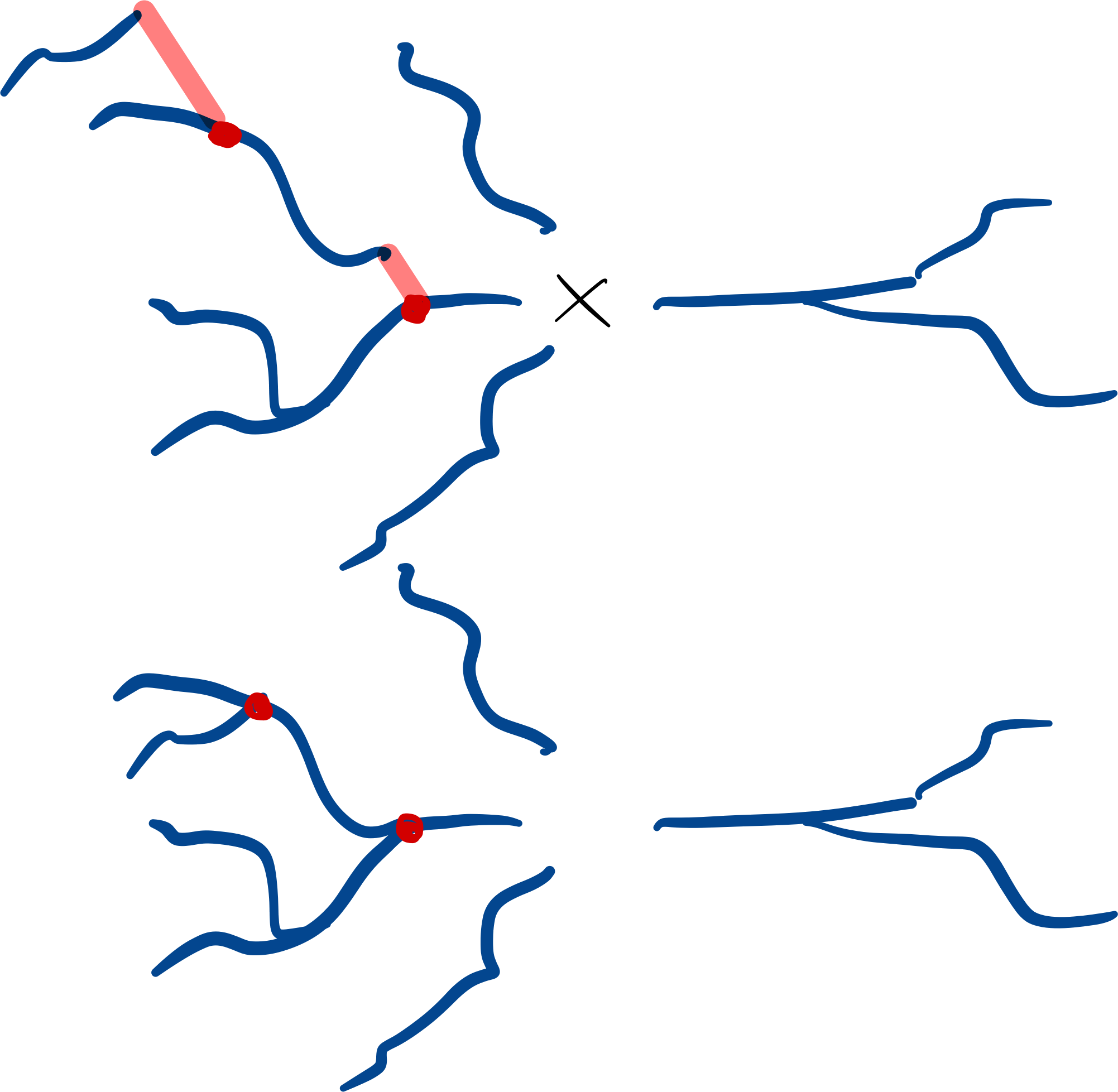
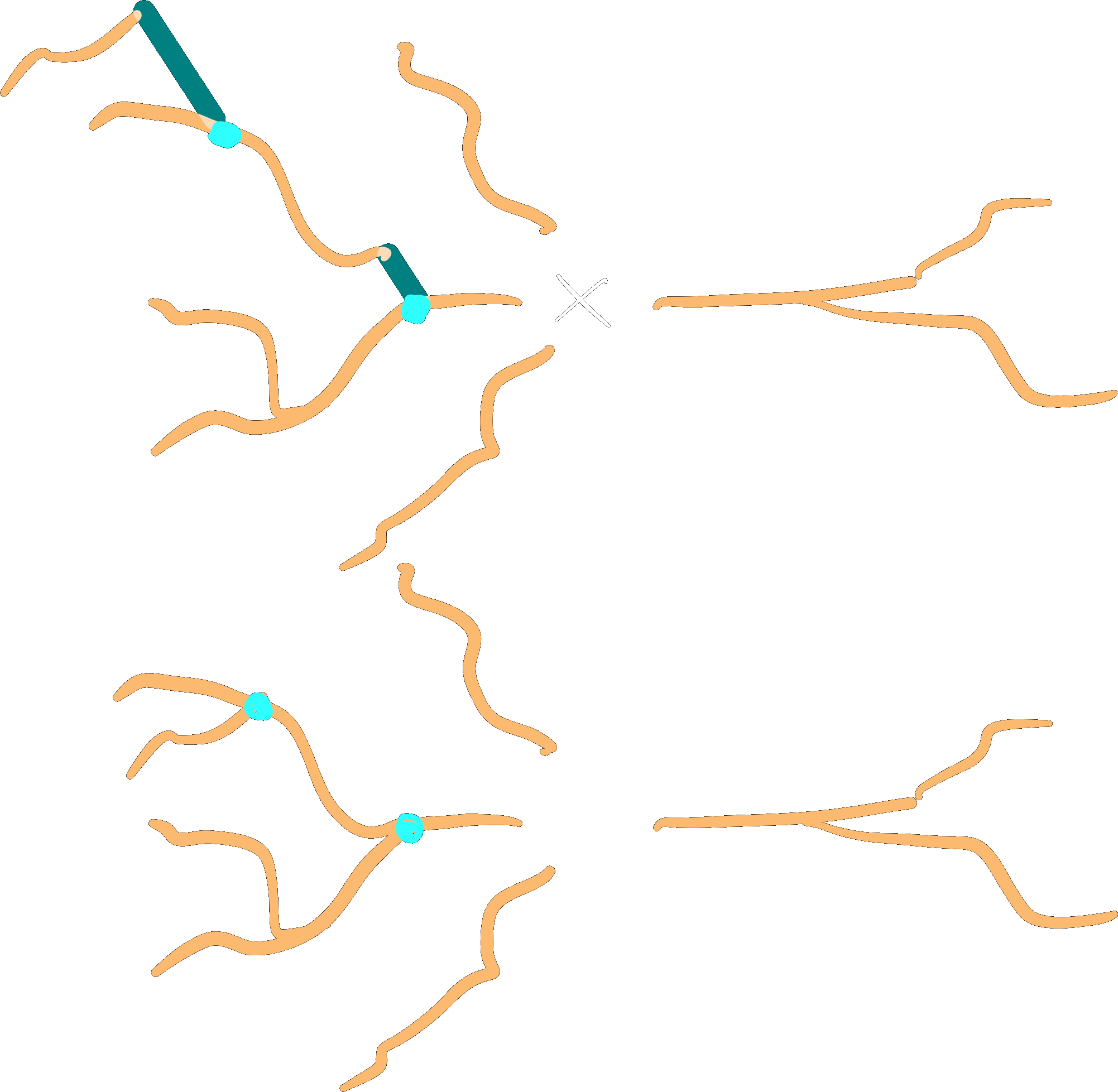
dendrites.close_gaps()
Note
The gaps between any subtree branch and its parent will be closed, even if the parent is not part of the subtree. This means that gaps of roots of a subtree may be closed as well. Gaps _between_ roots are never collapsed.
See also
Collapsing#
Collapse the roots of a subtree onto a single point, by default the origin.
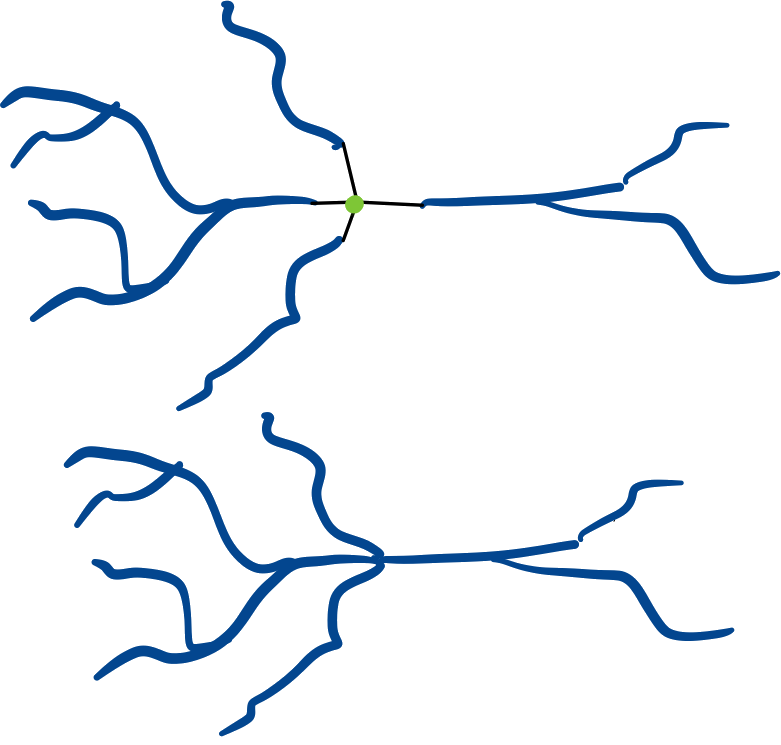
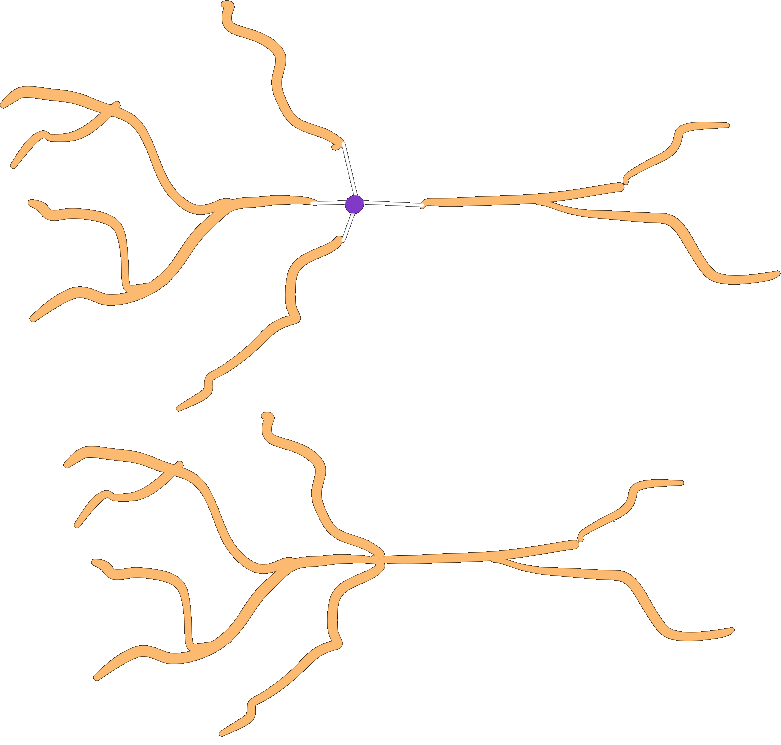
roots.collapse()
Call chaining
Calls to any of the above functions can be chained together:
dendrites.close_gaps().center().rotate(r)
Advanced features#
Morphology preloading#
Reading the morphology data from the repository takes time. Usually morphologies are
passed around in the framework as StoredMorphologies. These objects have a
load() method to load the
Morphology object from storage and a
get_meta() method to return the metadata.
Morphology selectors#
The most common way of telling the framework which morphologies to use is through
MorphologySelectors. Currently you
can select morphologies by_name or from_neuromorpho:
"morphologies": [
{
"select": "by_name",
"names": ["my_morpho_1", "all_other_*"]
},
{
"select": "from_neuromorpho",
"names": ["H17-03-013-11-08-04_692297214_m", "cell010_GroundTruth"]
}
]
If you want to make your own selector, you should implement the
validate() and
pick() methods.
validate can be used to assert that all the required morphologies and metadata are
present, while pick needs to return True/False to include a morphology in the
selection. Both methods are handed StoredMorphology objects.
Only load() morphologies if it is impossible
to determine the outcome from the metadata alone.
The following example creates a morphology selector selects morphologies based on the
presence of a user defined metadata "size":
from bsb import config, MorphologySelector
@config.node
class MySizeSelector(MorphologySelector, classmap_entry="by_size"):
min_size = config.attr(type=float, default=20)
max_size = config.attr(type=float, default=50)
def validate(self, morphos):
if not all("size" in m.get_meta() for m in morphos):
raise Exception("Missing size metadata for the size selector")
def pick(self, morpho):
meta = morpho.get_meta()
return meta["size"] > self.min_size and meta["size"] < self.max_size
After installing your morphology selector as a plugin, you can use by_size as
selector:
{
"cell_type_A": {
"spatial": {
"morphologies": [
{
"select": "by_size",
"min_size": 35
}
]
}
}
}
network.cell_types.cell_type_A.spatial.morphologies = [MySizeSelector(min_size=35)]
Morphology metadata#
Currently unspecified, up to the Storage and MorphologyRepository support to return a
dictionary of available metadata from
get_meta().
Morphology distributors#
A MorphologyDistributor is a special type of
Distributor that is called after positions have been
generated by a PlacementStrategy to assign morphologies, and
optionally rotations. The distribute()
method is called with the partitions, the indicators for the cell type and the positions;
the method has to return a MorphologySet or a tuple together with
a RotationSet.
Warning
The rotations returned by a morphology distributor may be overruled when a
RotationDistributor is defined for the same placement
block.
Distributor configuration#
Each placement block may contain a
DistributorsNode, which can specify the morphology and/or
rotation distributors, and any other property distributor:
{
"placement": {
"placement_A": {
"strategy": "bsb.placement.RandomPlacement",
"cell_types": ["cell_A"],
"partitions": ["layer_A"],
"distribute": {
"morphologies": {
"strategy": "roundrobin"
}
}
}
}
}
from bsb import RoundRobinMorphologies
network.placement.placement_A.distribute.morphologies = RoundRobinMorphologies()
Distributor interface#
The generic interface has a single function: distribute(positions, context). The
context contains .partitions and .indicator for additional placement context.
The distributor must return a dataset of N floats, where N is the number of positions
you’ve been given, so that it can be stored as an additional property on the cell type.
The morphology distributors have a slightly different interface, and receive an additional
morphologies argument: distribute(positions, morphologies, context). The
morphologies are a list of StoredMorphology, that the user
has configured to use for the cell type under consideration and that the distributor
should consider the input, or template morphologies for the operation.
The morphology distributor is supposed to return an array of N integers, where each
integer refers to an index in the list of morphologies. e.g.: if there are 3 morphologies,
putting a 0 on the n-th index means that cell N will be assigned morphology 0
(which is the first morphology in the list). 1 and 2 refer to the 2nd and 3rd
morphology, and returning any other values would be an error.
If you need to break out of the morphologies that were handed to you, morphology
distributors are also allowed to return their own MorphologySet.
Since you’re free to pass any list of morphology loaders to create a morphology set, you
can put and assign any morphology you like.
Tip
MorphologySets work on
StoredMorphologies! This means that it
is your job to save the morphologies into your network first, and to use the returned
values of the save operation as input to the morphology set:
def distribute(self, positions, morphologies, context):
# We're ignoring what is given, and make our own morphologies
morphologies = [Morphology(...) for p in positions]
# If we pass the `morphologies` to the `MorphologySet`, we create an error.
# So we save the morphologies, and use the stored morphologies instead.
loaders = [
self.scaffold.morphologies.save(f"morpho_{i}", m)
for i, m in enumerate(morphologies)
]
return MorphologySet(loaders, np.arange(len(loaders)))
This is cumbersome, so if you plan on generating new morphologies, use a morphology generator instead.
Finally, each morphology distributor is allowed to return an additional argument to assign
rotations to each cell as well. The return value must be a
RotationSet.
Warning
The rotations returned from a morphology distributor may be ignored and replaced by the values of the rotation distributor, if the user configures one.
The following example creates a distributor that selects smaller morphologies the closer the position is to the top of the partition:
import numpy as np
from scipy.stats.distributions import norm
from bsb import MorphologyDistributor
class SmallerTopMorphologies(MorphologyDistributor, classmap_entry="small_top"):
def distribute(self, positions, morphologies, context):
# Get the maximum Y coordinate of all the partitions boundaries
top_of_layers = np.maximum([p.data.mdc[1] for p in context.partitions])
depths = top_of_layers - positions[:, 1]
# Get all the heights of the morphologies, by peeking into the morphology metadata
msizes = [
loader.get_meta()["mdc"][1] - loader.get_meta()["ldc"][1]
for loader in morphologies
]
# Pick deeper positions for bigger morphologies.
weights = np.column_stack(
[norm(loc=size, scale=20).pdf(depths) for size in msizes]
)
# The columns are the morphology ids, so make an arr from 0 to n morphologies.
picker = np.arange(weights.shape[1])
# An array to store the picked weights
picked = np.empty(weights.shape[0], dtype=int)
rng = np.default_rng()
for i, p in enumerate(weights):
# Pick a value from 0 to n, based on the weights.
picked[i] = rng.choice(picker, p=p)
# Return the picked morphologies for each position.
return picked
Then, after installing your distributor as a plugin, you can use small_top:
{
"placement": {
"placement_A": {
"strategy": "bsb.placement.RandomPlacement",
"cell_types": ["cell_A"],
"partitions": ["layer_A"],
"distribute": {
"morphologies": {
"strategy": "small_top"
}
}
}
}
}
network.placement.placement_A.distribute.morphologies = SmallerTopMorphologies()
Morphology generators#
Continuing on the morphology distributor, one can also make a specialized generator of
morphologies. The generator takes the same arguments as a distributor, but returns a list
of Morphology objects, and the morphology indices to make use of
them. It can also return rotations as a 3rd return value.
This example is a morphology generator that generates a simple stick that drops down to the origin for each position:
import numpy as np
from bsb import Branch, Morphology, MorphologyGenerator
class TouchTheBottomMorphologies(MorphologyGenerator, classmap_entry="touchdown"):
def generate(self, positions, morphologies, context):
return [
Morphology([Branch([pos, [pos[1], 0, pos[2]]], [1, 1])]) for pos in positions
], np.arange(len(positions))
Then, after installing your generator as a plugin, you can use touchdown:
{
"placement": {
"placement_A": {
"strategy": "bsb.placement.RandomPlacement",
"cell_types": ["cell_A"],
"partitions": ["layer_A"],
"distribute": {
"morphologies": {
"strategy": "touchdown"
}
}
}
}
}
network.placement.placement_A.distribute.morphologies = TouchTheBottomMorphologies()
MorphologySets#
MorphologySets are the result of
distributors assigning morphologies
to placed cells. They consist of a list of StoredMorphologies, a vector of indices referring to these stored
morphologies and a vector of rotations. You can use
iter_morphologies() to iterate over each morphology.
ps = network.get_placement_set("my_detailed_neurons")
positions = ps.load_positions()
morphology_set = ps.load_morphologies()
rotations = ps.load_rotations()
cache = morphology_set.iter_morphologies(cache=True)
for pos, morpho, rot in zip(positions, cache, rotations):
morpho.rotate(rot)
Reference#
Morphology module
- class bsb.morphologies.Branch(points, radii, labels=None, properties=None, children=None)[source]#
A vector based representation of a series of point in space. Can be a root or connected to a parent branch. Can be a terminal branch or have multiple children.
- as_arc()[source]#
Return the branch as a vector of arclengths in the closed interval [0, 1]. An arclength is the distance each point to the start of the branch along the branch axis, normalized by total branch length. A point at the start will have an arclength close to 0, and a point near the end an arclength close to 1
- Returns:
Vector of branch points as arclengths.
- Return type:
- attach_child(branch)[source]#
Attach a branch as a child to this branch.
- Parameters:
branch (
Branch) – Child branch
- cached_voxelize(N)[source]#
Turn the morphology or subtree into an approximating set of axis-aligned cuboids and cache the result.
- Return type:
- center()#
Center the morphology on the origin
- property children#
Collection of the child branches of this branch.
- close_gaps()#
Close any head-to-tail gaps between parent and child branches.
- collapse(on=None)#
Collapse all the roots of the morphology or subtree onto a single point.
- Parameters:
on (int) – Index of the root to collapse on. Collapses onto the origin by default.
- contains_labels(labels)[source]#
Check if this branch contains any points labelled with any of the given labels.
- copy(branch_class=None)[source]#
Return a parentless and childless copy of the branch.
- Parameters:
branch_class (type) – Custom branch creation class
- Returns:
A branch, or branch_class if given, without parents or children.
- Return type:
- delete_point(index)[source]#
Remove a point from the branch
- Parameters:
index (int) – index position of the point to remove
- Returns:
the branch where the point has been removed
- Return type:
- detach_child(branch)[source]#
Remove a branch as a child from this branch.
- Parameters:
branch (
Branch) – Child branch
- property end#
Return the spatial coordinates of the terminal point of this branch.
- property euclidean_dist#
Return the Euclidean distance from the start to the terminal point of this branch.
- find_closest_point(coord)[source]#
Return the index of the closest on this branch to a desired coordinate.
- Parameters:
coord – The coordinate to find the nearest point to
- Type:
- flatten()#
Return the flattened points of the morphology or subtree.
- Return type:
- flatten_labels()#
Return the flattened labels of the morphology or subtree.
- Return type:
- flatten_properties()#
Return the flattened properties of the morphology or subtree.
- Return type:
- flatten_radii()#
Return the flattened radii of the morphology or subtree.
- Return type:
- property fractal_dim#
Return the fractal dimension of this branch, computed as the coefficient of the line fitting the log-log plot of path vs euclidean distances of its points.
- get_axial_distances(idx_start=0, idx_end=-1, return_max=False)[source]#
Return the displacements or its max value of a subset of branch points from its axis vector. :param idx_start = 0: index of the first point of the subset. :param idx_end = -1: index of the last point of the subset. :param return_max = False: if True the function only returns the max value of displacements, otherwise the entire array.
- get_branches(labels=None)#
Return a depth-first flattened array of all or the selected branches.
- get_label_mask(labels)[source]#
Return a mask for the specified labels
- Parameters:
labels (List[str] | numpy.ndarray[str]) – The labels to check for.
- Returns:
A boolean mask that selects out the points that match the label.
- Return type:
List[numpy.ndarray]
- get_points_labelled(labels)[source]#
Filter out all points with certain labels
- Parameters:
labels (List[str] | numpy.ndarray[str]) – The labels to check for.
- Returns:
All points with the labels.
- Return type:
List[numpy.ndarray]
- insert_branch(branch, index)[source]#
Split this branch and insert the given
branchat the specifiedindex.- Parameters:
branch (
Branch) – Branch to be attachedindex – Index or coordinates of the cutpoint; if coordinates are given, the closest point to the coordinates is used.
- Type:
Union[
numpy.ndarray, int]
- introduce_point(index, *args, labels=None)[source]#
Insert a new point at
index, before the existing point atindex.
- property is_root#
Returns whether this branch is root or if it has a parent.
- Returns:
True if this branch has no parent, False otherwise.
- Return type:
- property is_terminal#
Returns whether this branch is terminal or if it has children.
- Returns:
True if this branch has no children, False otherwise.
- Return type:
- label(labels, points=None)[source]#
Add labels to the branch.
- Parameters:
labels (List[str]) – Label(s) for the branch
points – An integer or boolean mask to select the points to label.
- property labels#
Return the labels of the points on this branch. Labels are represented as a number that is associated to a set of labels. See Labels for more info.
- property labelsets#
Return the sets of labels associated to each numerical label.
- property max_displacement#
Return the max displacement of the branch points from its axis vector.
- property path_length#
Return the sum of the euclidean distances between the points on the branch.
- property point_vectors#
Return the individual vectors between consecutive points on this branch.
- property points#
Return the spatial coordinates of the points on this branch.
- property radii#
Return the radii of the points on this branch.
- root_rotate(rot, downstream_of=0)#
Rotate the subtree emanating from each root around the start of that root If downstream_of is provided, will rotate points starting from the index provided (only for subtrees with a single root).
- Parameters:
rot (scipy.spatial.transform.Rotation) – Scipy rotation to apply to the subtree.
downstream_of – index of the point in the subtree from which the rotation should be applied. This feature works only when the subtree has only one root branch.
- Returns:
rotated Morphology
- Return type:
- rotate(rotation, center=None)#
Point rotation
- Parameters:
rot – Scipy rotation
center (numpy.ndarray) – rotation offset point.
- Type:
Union[scipy.spatial.transform.Rotation, List[float,float,float]]
- property segments#
Return the start and end points of vectors between consecutive points on this branch.
- simplify(epsilon, idx_start=0, idx_end=-1)[source]#
Apply Ramer–Douglas–Peucker algorithm to all points or a subset of points of the branch. :param epsilon: Epsilon to be used in the algorithm. :param idx_start = 0: Index of the first element of the subset of points to be reduced. :param epsilon = -1: Index of the last element of the subset of points to be reduced.
- simplify_branches(epsilon)#
Apply Ramer–Douglas–Peucker algorithm to all points of all branches of the SubTree. :param epsilon: Epsilon to be used in the algorithm.
- property size#
Returns the amount of points on this branch
- Returns:
Number of points on the branch.
- Return type:
- property start#
Return the spatial coordinates of the starting point of this branch.
- translate(point)#
Translate the subtree by a 3D vector.
- Parameters:
point (numpy.ndarray) – 3D vector to translate the subtree.
- Returns:
the translated subtree
- Return type:
- property vector#
Return the vector of the axis connecting the start and terminal points.
- property versor#
Return the normalized vector of the axis connecting the start and terminal points.
- voxelize(N)#
Turn the morphology or subtree into an approximating set of axis-aligned cuboids.
- Return type:
- class bsb.morphologies.Morphology(roots, meta=None, shared_buffers=None, sanitize=False)[source]#
A multicompartmental spatial representation of a cell based on a directed acyclic graph of branches whom consist of data vectors, each element of a vector being a coordinate or other associated data of a point on the branch.
- property adjacency_dictionary#
Return a dictonary associating to each key (branch index) a list of adjacent branch indices
- as_filtered(labels=None)[source]#
Return a filtered copy of the morphology that includes only points that match the current label filter, or the specified labels.
- get_label_mask(labels)[source]#
Get a mask corresponding to all the points labelled with 1 or more of the given labels
- property labelsets#
Return the sets of labels associated to each numerical label.
- set_label_filter(labels)[source]#
Set a label filter, so that as_filtered returns copies filtered by these labels.
- class bsb.morphologies.MorphologySet(loaders, m_indices=None, /, labels=None)[source]#
Associates a set of
StoredMorphologiesto cells- iter_morphologies(cache=True, unique=False, hard_cache=False)[source]#
Iterate over the morphologies in a MorphologySet with full control over caching.
- Parameters:
cache (bool) – Use Soft caching (1 copy stored in mem per cache miss, 1 copy created from that per cache hit).
hard_cache – Use Soft caching (1 copy stored on the loader, always same copy returned from that loader forever).
- class bsb.morphologies.RotationSet(data)[source]#
Set of rotations. Returned rotations are of
scipy.spatial.transform.Rotation
- class bsb.morphologies.SubTree(branches, sanitize=True)[source]#
Collection of branches, not necesarily all connected.
- property branch_adjacency#
Return a dictonary containing mapping the id of the branch to its children.
- property branches#
Return a depth-first flattened array of all branches.
- cached_voxelize(N)[source]#
Turn the morphology or subtree into an approximating set of axis-aligned cuboids and cache the result.
- Return type:
- collapse(on=None)[source]#
Collapse all the roots of the morphology or subtree onto a single point.
- Parameters:
on (int) – Index of the root to collapse on. Collapses onto the origin by default.
- flatten_properties()[source]#
Return the flattened properties of the morphology or subtree.
- Return type:
- get_branches(labels=None)[source]#
Return a depth-first flattened array of all or the selected branches.
- label(labels, points=None)[source]#
Add labels to the morphology or subtree.
- Parameters:
points (numpy.ndarray) – Optional boolean or integer mask for the points to be labelled.
- property path_length#
Return the total path length as the sum of the euclidian distances between consecutive points.
- root_rotate(rot, downstream_of=0)[source]#
Rotate the subtree emanating from each root around the start of that root If downstream_of is provided, will rotate points starting from the index provided (only for subtrees with a single root).
- Parameters:
rot (scipy.spatial.transform.Rotation) – Scipy rotation to apply to the subtree.
downstream_of – index of the point in the subtree from which the rotation should be applied. This feature works only when the subtree has only one root branch.
- Returns:
rotated Morphology
- Return type:
- rotate(rotation, center=None)[source]#
Point rotation
- Parameters:
rot – Scipy rotation
center (numpy.ndarray) – rotation offset point.
- Type:
Union[scipy.spatial.transform.Rotation, List[float,float,float]]
- simplify_branches(epsilon)[source]#
Apply Ramer–Douglas–Peucker algorithm to all points of all branches of the SubTree. :param epsilon: Epsilon to be used in the algorithm.
- translate(point)[source]#
Translate the subtree by a 3D vector.
- Parameters:
point (numpy.ndarray) – 3D vector to translate the subtree.
- Returns:
the translated subtree
- Return type: